Introduction
•
A diffusion model, fine-tuned on the RoseTTAFold2 (RF2)
•
Capable of generating protein backbones on various conditions (unconditional, topology-constrained monomer, binder, symmetric oligomer, motif scaffolding, metal-binding property)
•
Also backed up with various experimental results and structures of the generated backbones.
DDPMs (Denoising diffusion probabilistic models)
•
DDPMs have shown powerful performance on generating realistic images (with & without conditions)
Pros in applying DDPMs in protein design:
•
Diversity
•
Easy to provide guidance
•
Can explicitly model the equivariance in protein 3D structures
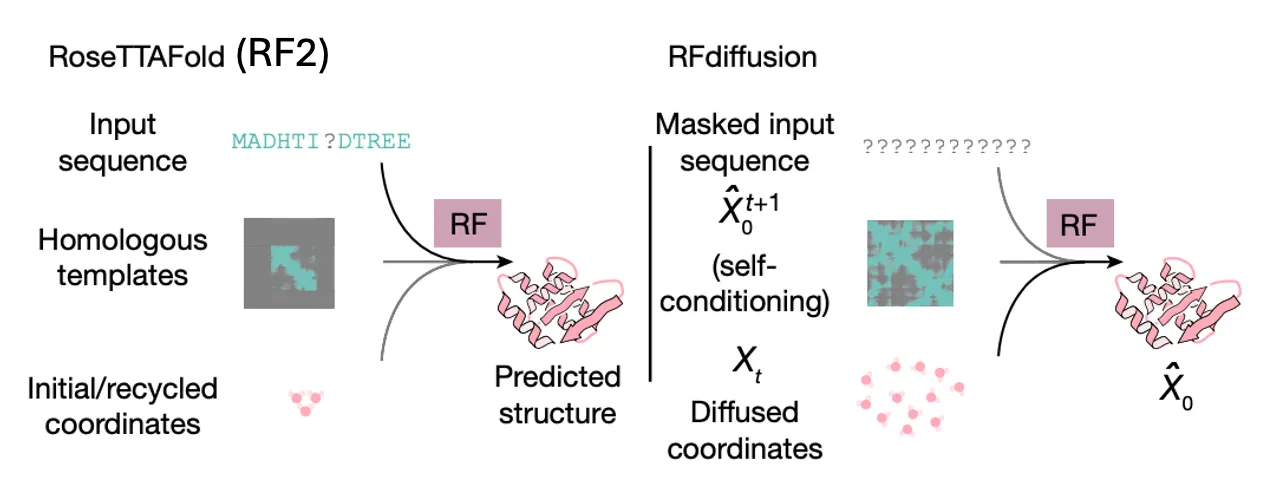
RF2 (RoseTTAFold2): A structure prediction model (base pretrained model for RFdiffusion)
•
Background:
RF1 (2021, a structure prediction model) was developed first.
RF2 (2023, also a structure prediction model) came out later, with significant architectural & loss modification.
Suppl. Fig 1. The three-track (1D, 2D, and 3D feature track) architecture of RF2.
Notable architectural differences compared to RF1
•
RF2 is the base (pretrained model) of RFdiffusion.
RFdiffusion: A backbone structure generation model 
•
Residue representation:
Frame same as RF, which is [ coordinate + rigid orientation]
•
Data: Structure data from PDB
•
Noise scheme:
◦
translation: perturb with 3D Gaussian noise
◦
residue orientation: Brownian motion on the manifold of rotation matrices
•
Loss:
MSE (mean-squared error) loss between frame predictions and the true protein structure (without alignment!)
Why MSE? Why not FAPE?
Background:
•
Originally, AlphaFold2 (2021, a structure prediction model) used FAPE loss.
•
FAPE loss is SE(3)-invariant.
•
RF1 (2021, a structure prediction model) did not use FAPE loss.
•
RF2 (2023, also a structure prediction model, and the base model of RFdiffusion) did use FAPE loss.
However, during RFdiffusion training, the input and the output of the model should be in the same global frame. For this purpose, MSE loss is much suitable than the FAPE loss.
•
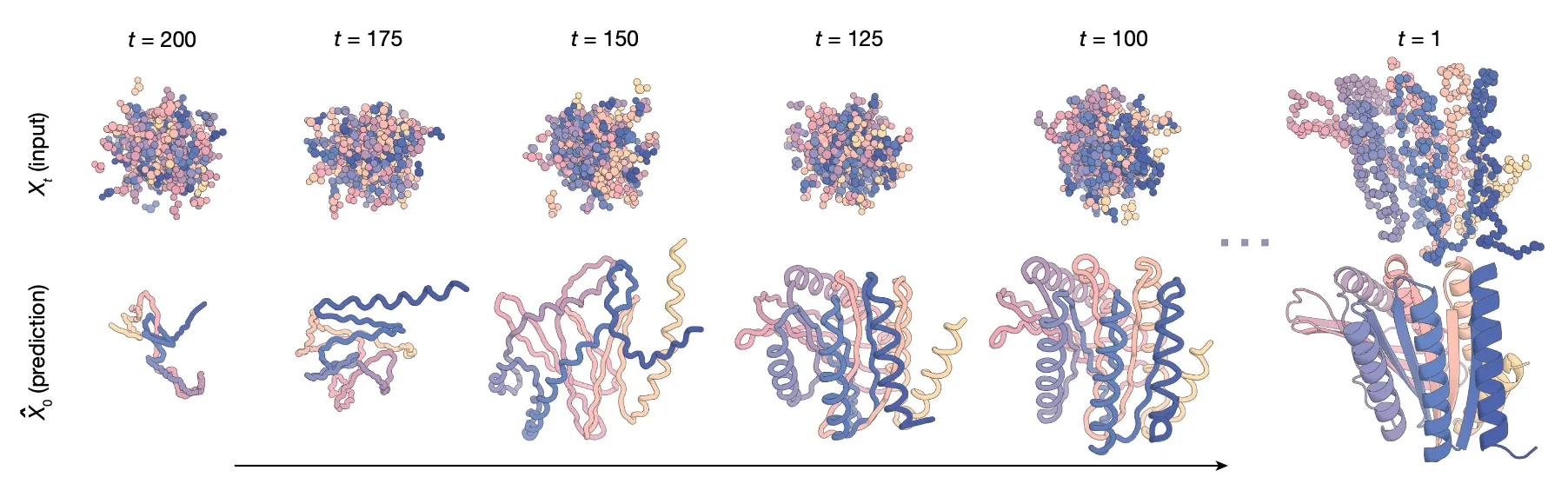
Structure Design:
Trajectory example (unconditional):
1.
First initialize random residue frames as and RFdiffusion makes a denoised prediction .
2.
Then the next input is set as the proper interpolation between and , and the RFdiffusion makes another denoised prediction .
3.
This goes on and on until the is 1.
•
Sequence Design:
Authors used ProteinMPNN network (a sequence design model).
The input of the ProteinMPNN network is the backbone structure (which is the output of the RFdiffusion model).
•
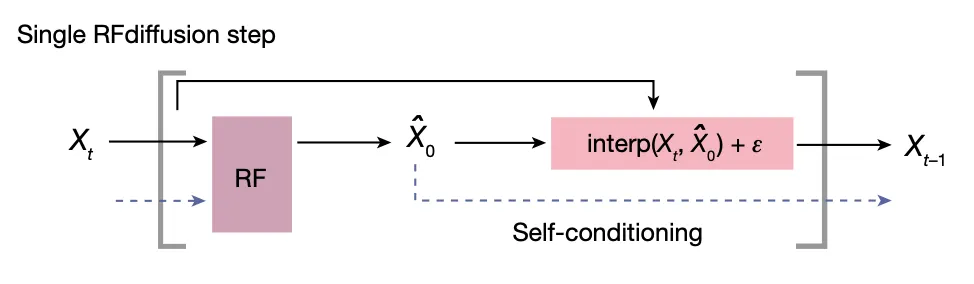
Notable training strategies
◦
Self-conditioning diffusion training scheme
The prediction from previous timestep is also provided to the model as template.
Why self-conditioning? Why not canonical (or typical) training?
Background:
•
Authors also tried canonical diffusion training scheme (which is the predictions at each timestep is independent of predictions at previous timesteps)
But authors found out that the self-conditioning notably improved performance.
◦
Fine-tuning from RF2
Authors found out fine-tuning RFdiffusion from pretrained RF2 weights was far more successful.
They used somewhat different inputs for each period.
RF2 (pre-training) | RFdiffusion (fine-tuning) | |
Sequence | input sequence
(some residues can be masked) | input sequence
(20~100% of the residues can be masked) |
Template (hint) | homologous template | coordinates to self-condition |
Input coordinate | initial or recycled coordinates | diffused coordinates (interpolated) |
•
Metrics for in silico ‘success’
“Success” when the AF2 predictions (from the RFdiffusion → ProteinMPNN sequence output) fulfills…
i.
High confidence of pAE (predicted aligned error) < 5
ii.
Globally within a 2Å backbone RMSD of the designed structure
iii.
Locally within a 1Å backbone RMSD on any scaffolded functional site
Authors reported that this measure correlates better with experimental success than TM-score based metrics.
Tasks
1. Unconditional & conditional protein monomer generation
Fig 2a.
Task explained:
Design a protein monomer with & without condition.
Difficulties of previous physics-based protein design methods:
Unconstrained generation of diverse monomers.
Because it was very hard to sample from extremely large and rugged conformational landscape.
RFdiffusion generates new structures!
It can generate plausible monomers with little similarity to training data.
Fig 2b. Highest TM score to known PDB structures decrease with the number of residues specified for generation → unconditional designs from RFdiffusion are new.
Fig 2c. Unconditional samples are closely repredicted by AF2 (up to 400 residues).
RFdiffusion can be guided with specific fold condition.
fine-tuned to condition on secondary structure and/or fold information.
Fig 2g. An RFdiffusion-generated sample, conditioned on the secondary structure and block adjacency of the PDB 6WVS structure.
2. Higher-order oligomer generation
Fig 3a. RFdiffusion-generated assemblies (overlaid with AF2 predictions, which shows little difference).
Fig 3b. The nsEM result of the designed assemblies.
Task explained:
Design symmetric oligomers.
Symmetric oligomers are important in bio-medical fields.
They can serve as vaccine platforms, delivery vehicles, and catalysts.
RFdiffusion can generate oligomers with cyclic (C3, C6, …), dihedral (D3, D4), tetrahedral, octahedral, and icosahedral(!) symmetries.
Symmetry is ensured through…
1.
Equivariant property (rotation)
2.
Explicit -copies of symmetrical starting point
3.
Explicit resymmetrization during each step
Generated oligomers were experimentally validated.
Out of 608 designs selected for experiments, at least 87 matched the intended oligomeric states based on SEC (size-exclusion chromatography)
3. Functional-motif scaffolding
Task explained:
Design the remaining scaffold, while maintaining the input motif coordinates.
25 motif-scaffolding design benchmarks
Authors established 25 benchmark tests from six recent publications.
RFdiffusion performs better than previous methods.
RFdiffusion solved 23 of the 25 benchmarks.
Fig 4a. RFdiffusion performs better than other methods across 25 benchmark problems.
RFdiffusion successfully generated the remaining scaffold, maintaining the input motif.
Fig 4b. Teal: input motif; colors: AF2 prediction of the design by RFdiffusion
4. Scaffolding enzyme active sites
Task explained:
Design the enzyme active sites that maintains a few single amino acid inputs
RFdiffusion can scaffold enzyme active sites (after some fine-tuning).
Fig 4f. With three amino acids as the input enzymatic sites, RFdiffusion can generate scaffolds!
5. Symmetric functional-motif scaffolding
Task explained:
Design symmetric protein assemblies that precisely position functional motifs.
RFdiffusion generated C3-symmetric timers that match the ACE2 binding sites on the SARS-CoV-2 spike protein trimer.
Fig 5a. RFdiffusion designed a C3-symmetric oligomer, scaffolding the binding interface of ACE2 mimic AHB2 against the SARS-CoV-2 spike trimer.
RFdiffusion generated C4-symmetric assemblies with histidine residues arranged in square-planar geometries for Ni2+ ion coordination.
Fig 5b. RFdiffusion designed a C4-symmetric oligomer to scaffold a Ni2+ binding motif.
6. Protein-binding protein generation 
Task explained:
Design a high-affinity binders that binds to specific regions of a target protein (with or without extra scaffold topology condition)
RFdiffusion was fine-tuned in the following two ways:
1.
With “interface hotspots” (as extra feature, which acts as condition)
To guide the model to generate some backbones to bind to specific region.
Fig 6a. RFdiffusion generates protein binders given a target and the interface hotspot residues.
Extended data Fig 8a. IL-7R has two patches for suitable binding.
Extended data Fig 8b. Without hotspot guidance, RFdiffusion almost always targeted the Site 1 (not Site 2). Site 1 is much more preferred with hotspot guidance, and even the binders for Site 2 can be generated with hotspot guidance.
2.
With “interface hotspots” + secondary structure info + block-adjacency info (also as extra feature and condition)
To guide the model even further to have some particular binder fold.
Extended data Fig 8c. RFdiffusion can also condition on the input fold information (secondary structure and block adjacency information)
In silico design with experimental validation
Five targets: Influenza A H1 Hemagglutinin (HA), Interleukin-7 Receptor-α (IL-7Rα), Programmed Death-Ligand 1 (PD-L1), Insulin Receptor (InsR), and Tropomyosin Receptor Kinase A (TrkA)
RFdiffusion achieved an experimental success rate of 19%.
This record is two-order-of-magnitude improvement over previous Rosetta-based method.
Fig 6b. RFdiffusion followed by AF2 filtering showed high experimental success rates.