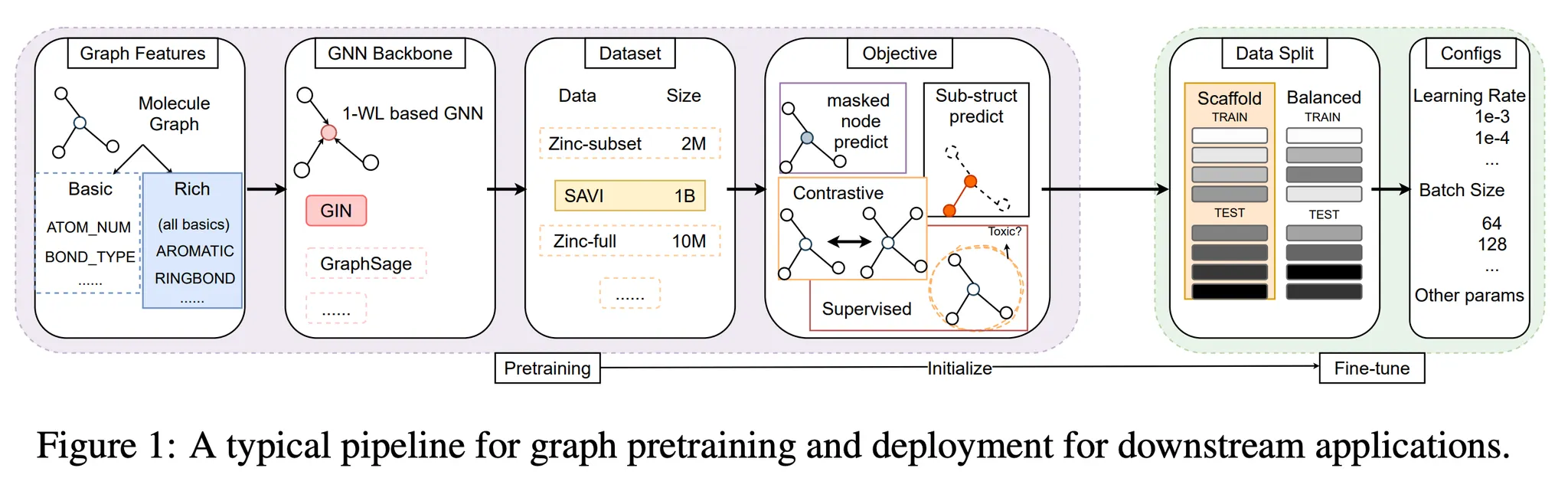
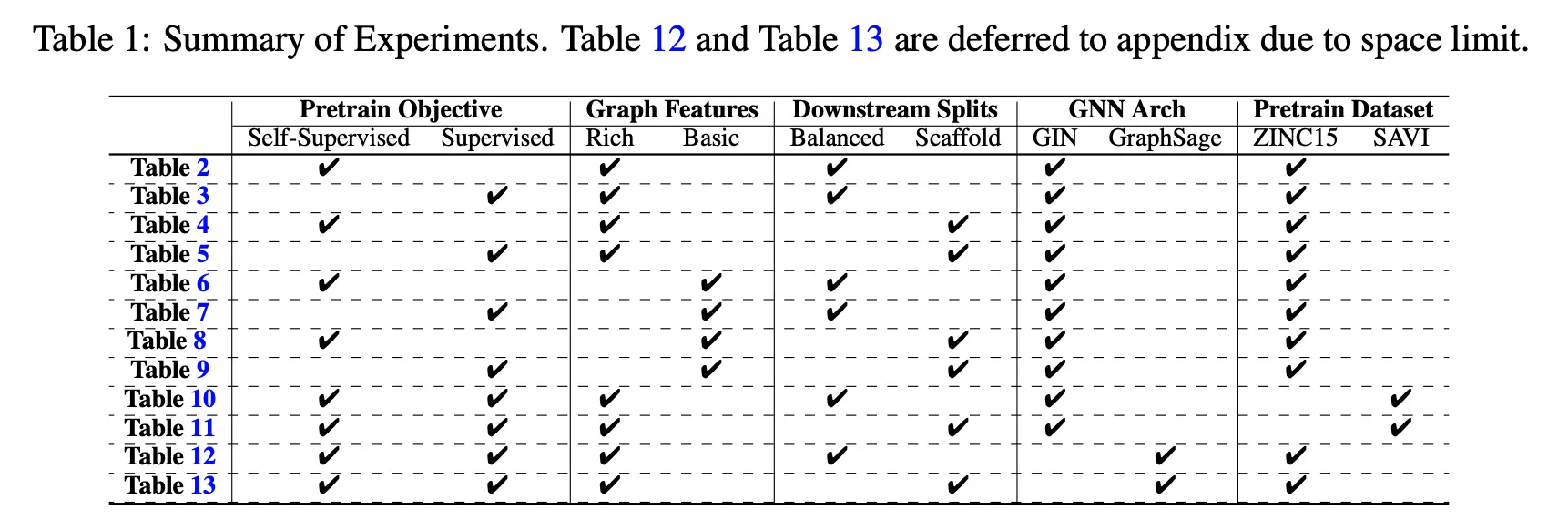
Summary
•
Self-supervised pre-training alone does not provide statistically significant improvements over non-pre-trained methods on downstream tasks.
•
Data splits, hand-crafted rich features, or hyperparameters can bring significant improvements.
Preliminaries
Pre-train objectives
•
Self-supervised
◦
Node prediction
◦
Context prediction
◦
Motif prediction
◦
Contrastive learning
•
Supervised
◦
Related tasks with label (e.g. ChEMBL dataset)
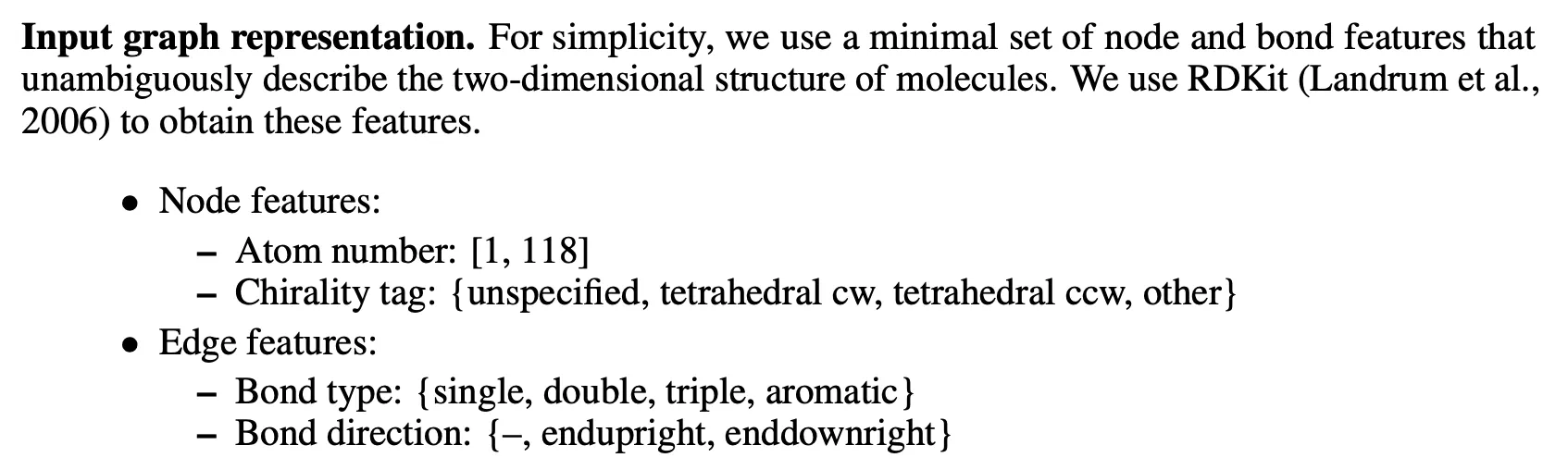
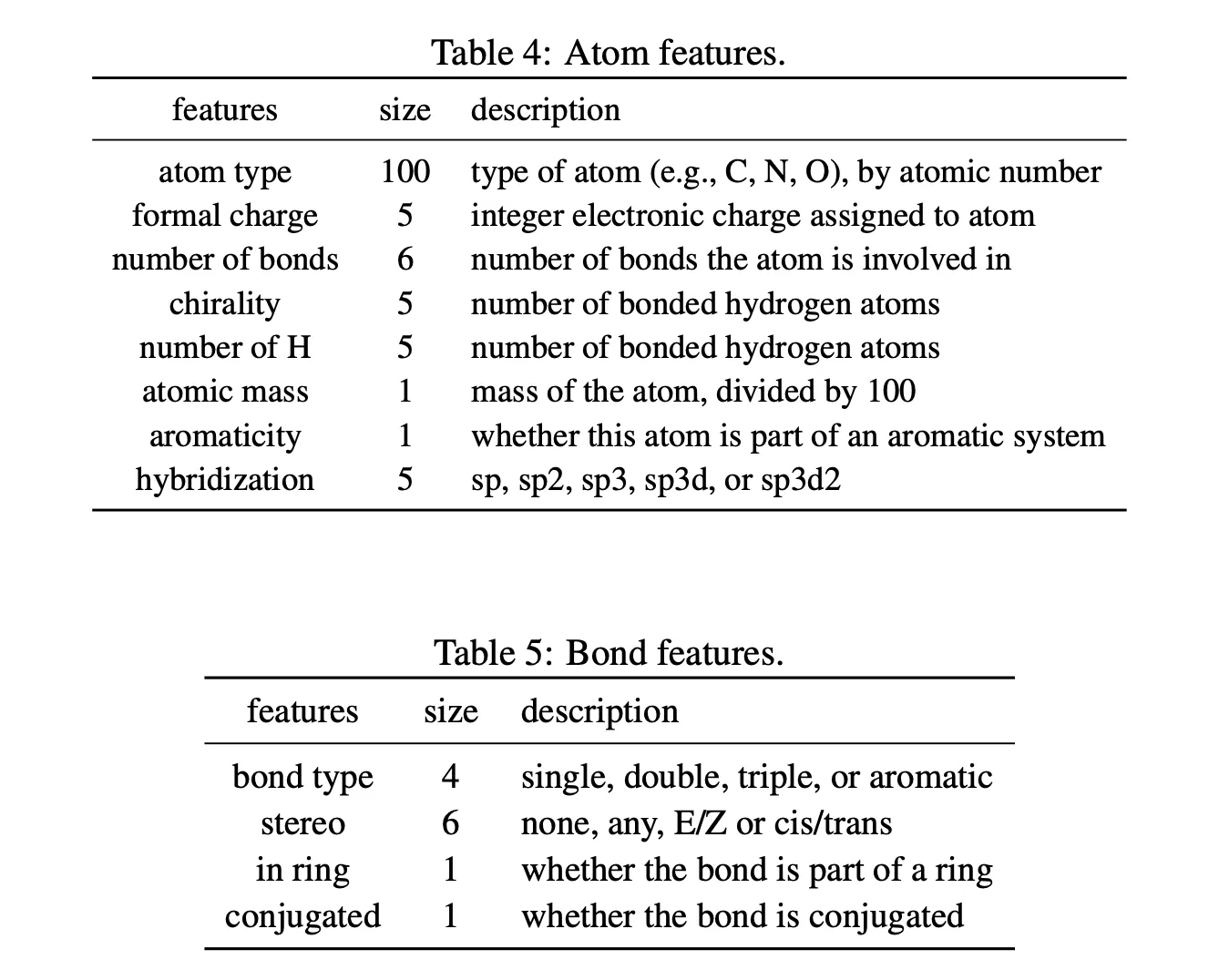
Graph features
•
•
Rich
In downstream tasks, additional 2d normalized rdNormalizedDescriptors are used (not in pre-training).
Downstream splits
•
Scaffold
Sorts the molecule according to the scaffold, then partition the sorted list into train/valid/test splits. → Deterministic
Molecules of each set are most different ones.
•
Balanced scaffold
Introduces the randomness in the sorting and splitting stages of Scaffold split.
GNN architecture
•
GIN
•
GraphSAGE
Pre-train dataset
•
ZINC15 (self-supervised)
2 million molecules. Pre-processed following Hu et al.
•
SAVI (self-supervised)
1 billion drug-like molecules synthesized by computer simulated reactions.
•
ChEMBL (supervised)
500k drugable molecules with 1,310 prediction target labels from bio-activity assays.
Results
Results tables
Key takeaways
When pre-training might help?
1.
Related supervised pre-training dataset. But not always feasible.
2.
If the rich features are absent.
3.
If the downstream split distributions are substantially different.
When the gain diminishes?
1.
If using rich features.
2.
If don’t have the highly relevant supervisions.
3.
If the downstream split is balanced.
4.
If the self-supervised learning dataset lacks diversity.
Why pre-training may not help in some cases?
Some of the pre-training methods (e.g. node label prediction) might be too easy
→ Transfer less knowledge.
Conclusion
1.
Use rich features
2.
Use balanced scaffold
3.
Use related supervised pre-training dataset
4.
Use difficult pre-training task (for self-supervised pre-training) and use high-quality negative samples.