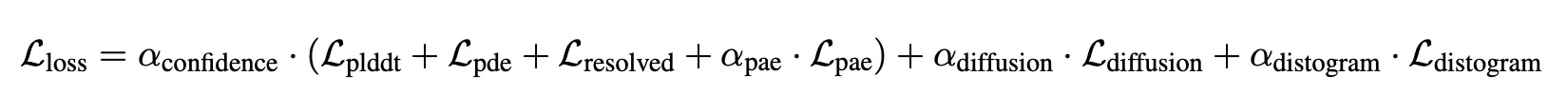Introduction
If you need some background information about protein folding, refer to  Introduction to Protein Folding
Introduction to Protein Folding 
Timeline of AlphaFold
•
2016
◦
AlphaGo beat Lee Sedol
◦
DeepMind hired a handful of biologists
•
2018
◦
CASP13
◦
AlphaFold1
•
•
2021
◦
•
2023
◦
AlphaFold latest (blog post)
•
AlphaFold3
An almighty AF model for nearly all bio-molecular types
Major differences with AF2
Data type & processing
AF2 | AF3 | |
Data type | Protein (polypeptide) only | General molecules (protein, nucleic acids, small molecules, ions, …) |
Tokenization | Amino acid residue frame & angles (for side chain atoms) | Atoms & tokens |
In AF3, atoms are grouped into tokens.
•
Standard amino acid & nucleic acid: token = entire nucleotides or residues
•
Others: token = a single heavy atom
Architecture
•
AF2 Evoformer | AF3 MSAModule | |
# blocks | 48 | 4 |
Sequence-level feature | MSA rep (row-wise & column-wise attention) | MSA rep (simple pair-weighted average) |
Structure-level feature | Pair rep (triangular multiplicative & self-attention updates) | Pair rep (triangular multiplicative & self-attention updates) |
Communication | Outer product mean, pair bias | Outer product mean, pair bias |
AF2 Evoformer | AF3 Pairformer | |
# blocks | 48 | 48 |
Sequence-level
feature | MSA rep (row-wise & column-wise attention) | Single rep (only row-wise attention) |
Structure-level
feature | Pair rep (triangular multiplicative & self-attention updates) | Pair rep (triangular multiplicative & self-attention updates) |
Communication | Outer product mean, pair bias | Pair bias |
In AF3, MSA representation is actually de-emphasized. It is first processed at the smaller and simpler MSAModule and then only the first row (single representation) is passed to the core Pairformer.
•
StructureModule → DiffusionModule
AF2 StructureModule | AF3 DiffusionModule | |
Model type | Predictive | Generative |
Resolution | Residue level | All-atom level |
Operates on | Residue frames and angles | Raw atom coordinates |
Equivariance | Forced to satisfy | Not required (!) |
Attention type | IPA (invariant point attention) | 1. Sequence local attention (atom-level)
2. Global attention (token-level)
3. Sequence local attention (atom-level) |
Major loss | FAPE loss | MSE loss (!) |
Initial coordinate | Black hole initialization | Generated conformer |
Confidence metrics
AF2 Confidence module | AF3 Confidence module | |
heads | - pAE head
- pLDDT head
- experimentally resolved head | - pAE head
- pLDDT head
- experimentally resolved head
- pDE head |
pAE head input | Pair rep (Evoformer output) | Pair rep (mini-Pairformer output - with input feature, single feature, distance between rolled-out coordinate information) |
pLDDT head input | Single rep (Structure module output) | Single rep (mini-Pairformer output - with input feature, pair feature, distance between rolled-out coordinate information) |
Input gradient | Not detached | Detached |
Activation function
•
ReLU → SwiGLU
Recycling scheme
AF2 | AF3 | |
4 | 4 | |
Recycled representation | - Single rep (Evoformer output)
- Pair rep (Evoformer output)
- Coordinate (StructureModule output) | - Single rep (Pairformer output)
- Pair rep (Pairformer output)
(coordinate is not recycled, but denoised at DiffusionModule) |
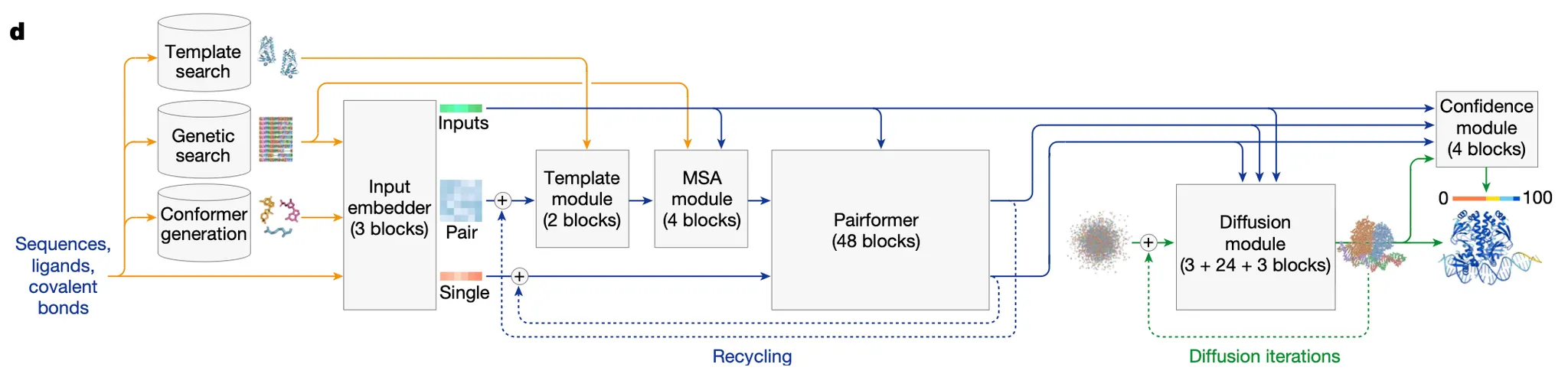
Architecture overview
c.f. AF2 architecture
Fig. 1d AF3 architecture for inference. yellow, input data; blue, abstract network activations; green, output data
•
Overall structure of AF3 resembles that of AF2.
◦
Large network trunk (Evoformer in AF2, Pairformer in AF3) evolves a pairwise representation.
◦
StructureModule (in AF2) or DiffusionModule (in AF3) uses the pairwise representation to generate explicit atomic positions.
•
Major modifications were made to..
1.
Accomodate a wide range of chemical entities without excessive special casing
2.
Increase the accuracy in protein structure predictions
•
AF3 is a conditional diffusion model, where most of the computation happens in the conditioning!
•
In AF3, coordinates are not recycled with Single rep or Pair rep. But they are iteratively denoised at the DiffusionModule.
Input processing
Tokenization
amino acid, nucleotide structure
Type | Tokenization | Token center atom |
Standard amino acid residue | Single token | CA |
Standard nucleotide residue | Single token | C1’ |
Modified amino acid / nucleotide | Tokenized per-atom | First (and only) atom |
All ligands | Tokenized per-atom | First (and only) atom |
Each token is designated with a token center atom.
Features
Type | AF2 | AF3 | Description |
MSA features | O | O | features from genetics search
(e.g. MSA itself, deletion matrix, profile) |
Template features | O | O | features from template search
(e.g. template atom positions) |
Token features | X | O | e.g. position indexes (token_index), chain identifiers (asym_id), masks (is_protein) |
Reference features | X | O | features from reference conformer (generated with RDKit ETKDGv3) |
Bond features | X | O | features about bond information |
TemplateModule (TemplateEmbedder)
Baby Pair representation builder 
Input: Template search info & (previous) Pair representation
Output: (updated) Pair representation
Goal: Build and update pair representation with the template information from the database
•
Template stack is similar to AF2.
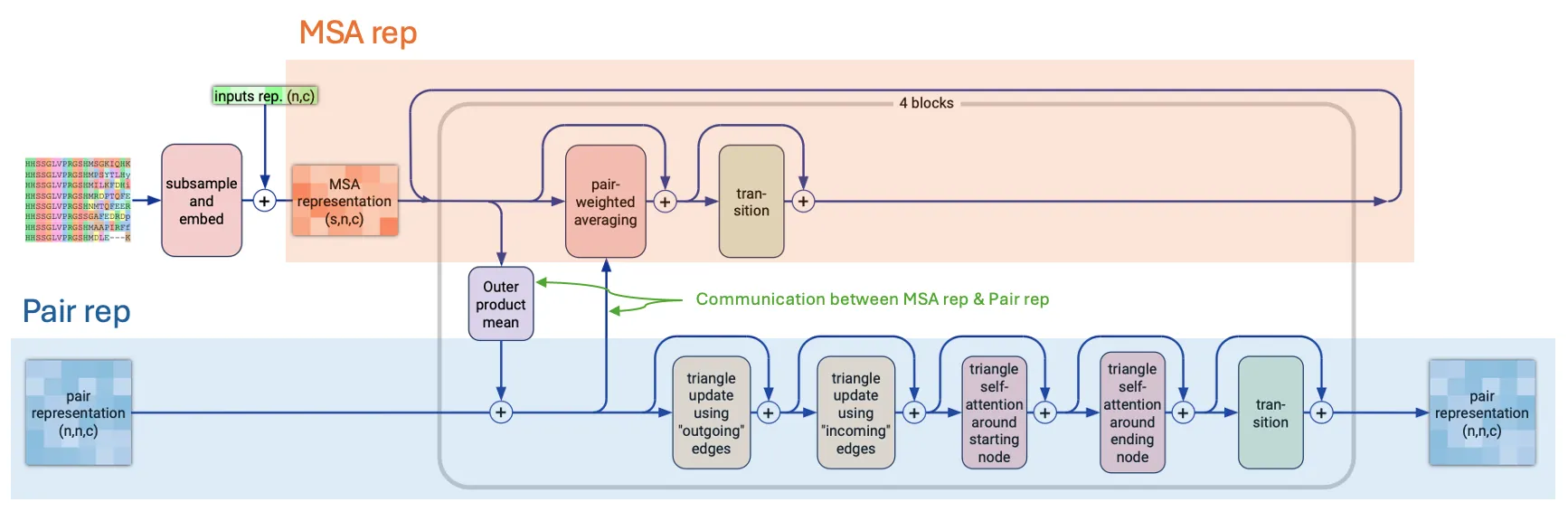
MSAModule
Mini-Evoformer 
Input: (previous) MSA representation & (previous) Pair representation
Output: (updated) MSA representation & (updated) Pair representation
Goal: Update MSA and pair representation while exchanging information
Suppl Fig. 2
•
MSAModule is practically a mini-Evoformer!
Authors divided Evoformer (of AF2) into MSAModule and Pairformer.
AF2 Evoformer | AF3 MSAModule | |
# blocks | 48 | 4 |
Sequence-level feature | MSA rep (row-wise & column-wise attention) | MSA rep (simple pair-weighted average) |
Structure-level feature | Pair rep (triangular multiplicative & self-attention updates) | Pair rep (triangular multiplicative & self-attention updates) |
Communication | Outer product mean, pair bias | Outer product mean, pair bias |
•
Motivation for this change:
1.
Enrich more information on Pair rep as it forms the backbone for the rest of the network!
2.
De-emphasize the MSA rep as there were some discussions that AF2 relies heavily on MSA.
•
In AF3, there is no key-query based attention in MSA stack and the architecture is similar to the ExtraMSAStack in AF2
→ Reduced computation & memory
•
MSA is used for protein and RNA sequences.
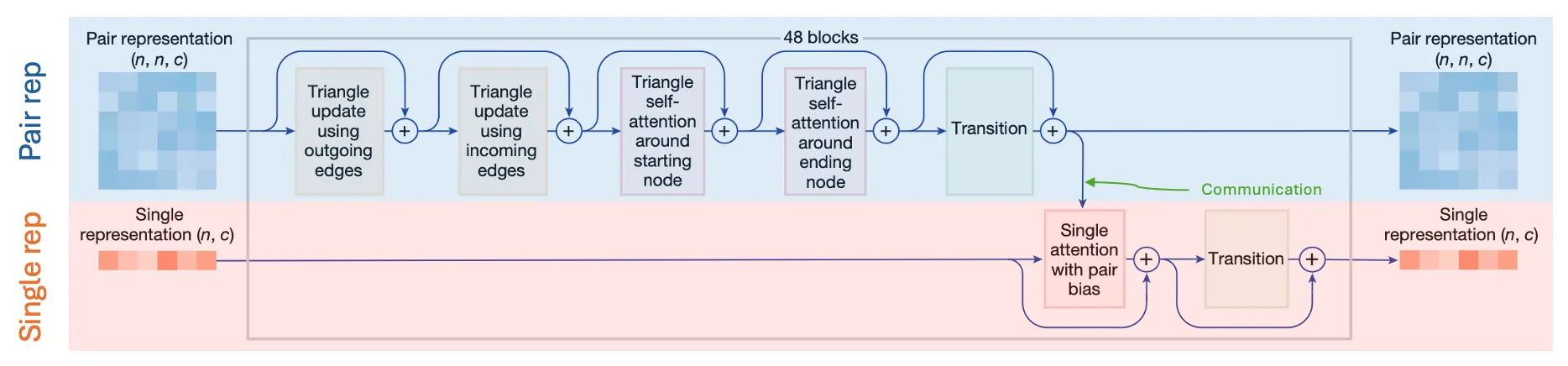
Pairformer 
The legitimate successor the the Evoformer 
Input: (previous) Single representation & (previous) Pair representation
Output: (updated) Single representation & (updated) Pair representation
Goal: Update single & pair representation while exchanging information
Fig. 2a Pairformer module. n: number of tokens, c: number of channels. Each 48 blocks does not share weights.
•
Pairformer is a light-weight Evoformer, focusing more on the Pair rep
AF2 Evoformer | AF3 Pairformer | |
# blocks | 48 | 48 |
Sequence-level
feature | MSA rep (row-wise & column-wise attention) | Single rep (only row-wise attention) |
Structure-level
feature | Pair rep (triangular multiplicative & self-attention updates) | Pair rep (triangular multiplicative & self-attention updates) |
Communication | Outer product mean, pair bias | Pair bias |
•
Single rep is the privileged first row of MSA rep.
•
Only row-wise attention for the Single rep!
In Pairformer, only row-wise attention is performed for the Single rep, since there is only one row (instead of rows in MSA rep)
•
No outer product mean in Pairformer!
Unlike in AF2, the Single rep does not (directly) influence the Pair rep within a Pairformer block with outer product mean.
(the outer product mean was already used in the MSA module)
DiffusionModule 
The rightful heir of the Structure module, without leash 
 and with modern generative technology
and with modern generative technology 
Input: Input feature, Single representation, Pair representation & (previous) Physical atom coordinates
Output: (updated) Physical atom coordinates
Goal: Update physical atom coordinates from original input feature and refined single & pair representation
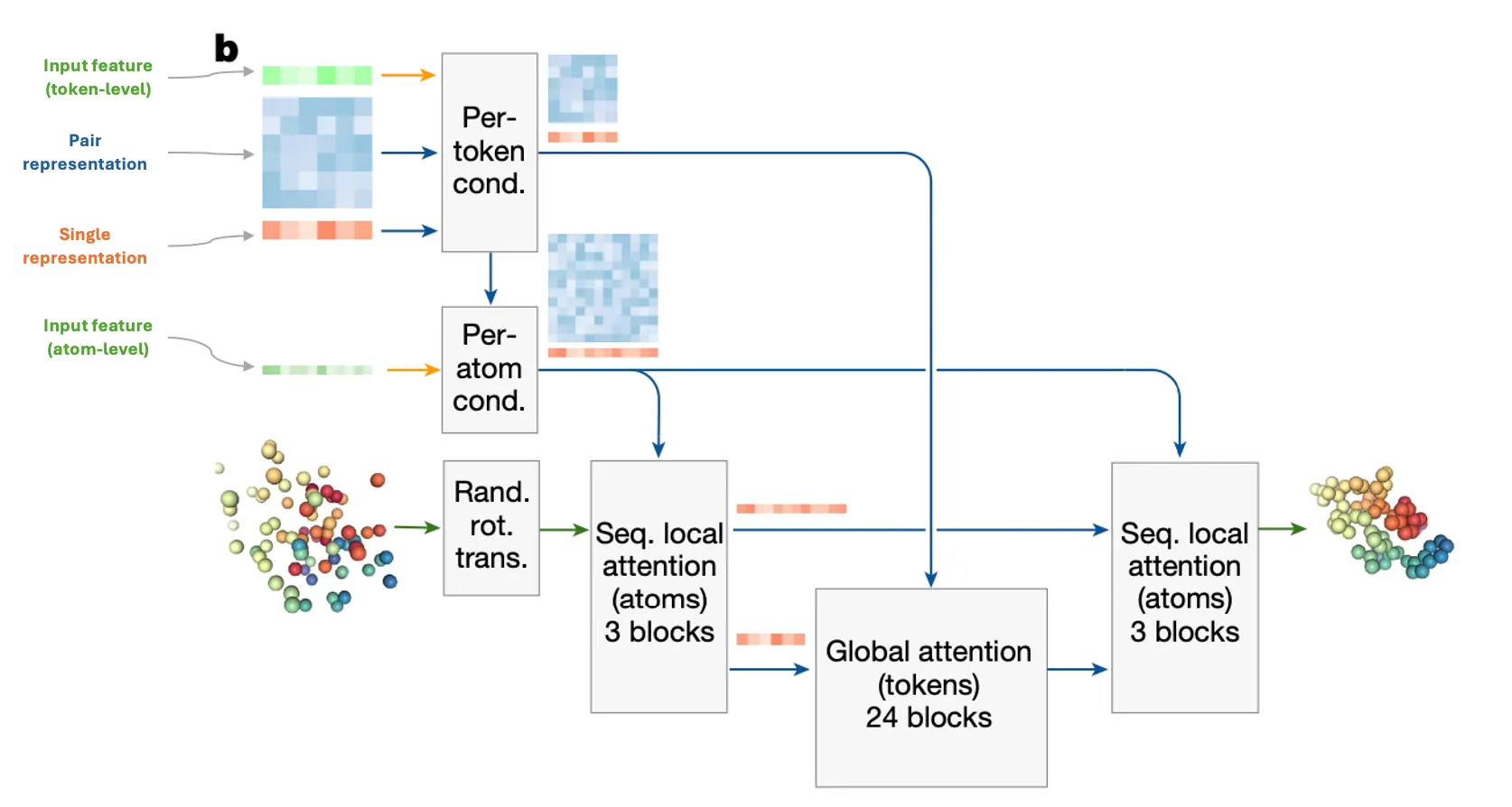
Fig. 2b Diffusion module. coarse: token, fine: atom. green: input, blue: pair, red: single.
•
DiffusionModule is an all-atom, generative, non-equivariant version of the StructureModule!
AF2 StructureModule | AF3 DiffusionModule | |
Model type | Predictive | Generative |
Resolution | Residue level | All-atom level |
Operates on | Residue frames and angles | Raw atom coordinates |
Equivariance | Forced to satisfy | Not required (!) |
Attention type | IPA (invariant point attention) | 1. Sequence local attention (atom-level)
2. Global attention (token-level)
3. Sequence local attention (atom-level) |
Major loss | FAPE loss | MSE loss (!) |
Initial coordinate | Black hole initialization | Generated conformer |
•
The diffusion module of AF3 is a conditional, non-equivariant generative model, that works on the point cloud of atoms.
◦
Condition: Input feature, Single rep, Pair rep
◦
Non-equivariance: no geometric inductive bias involved
In AF2, the Structure module had a invariant operation called IPA to fulfill equivariance constraints.
But in AF3, the authors found that no invariance or equivariance are required after all, and therefore omitted them to simplify the architecture.
•
Two-level architecture:
It works as follows: 1. on atom-level → 2. on token-level → 3. on atom-level
1. Sequence local attention on atom-level 
Allow nearby atoms to talk  directly to each other!
directly to each other!
Here, ‘nearby’ atoms are defined by sequence-level neighbors. (But why not structure-level neighbors?)
Each subset of 32 atoms attends to the subset of the nearby 128 atoms in sequence space.
This restriction is sub-optimal  , but was necessary to keep the memory and compute costs within reasonable bounds.
, but was necessary to keep the memory and compute costs within reasonable bounds.
2. Global attention on token-level
It’s time for the representatives (token) to gather local information and shake hands  with other global representatives!
with other global representatives!
1.
Aggregate fine-grained atom-level features into coarse-grained token-level features.
2.
Full self-attention on token-level.
3.
Broadcast token activations to atom features
3. Sequence local attention on atom-level AGAIN!
With that global information update, let’s focus on our local atom neighbors again.
DiffusionModule algorithm
Training setup
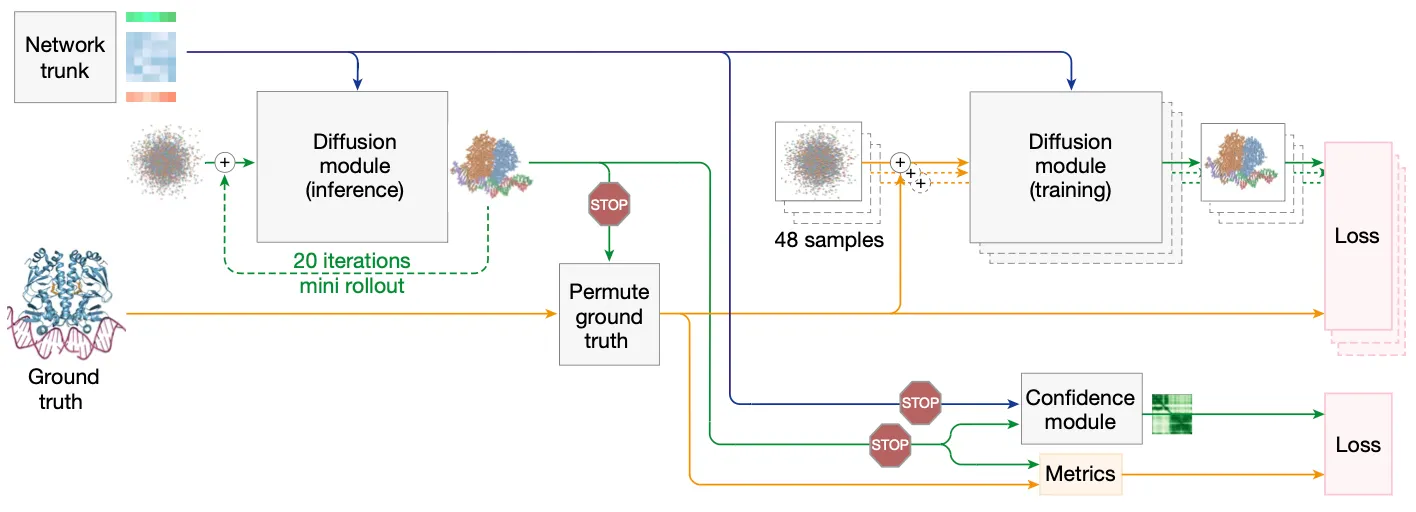
Fig. 2c Training setup (distogram head is omitted for simplicity) starting from the end of the network trunk. blue line: abstract activation, yellow line: ground truth, green line: predicted data
DiffusionModule Training
•
Efficient training of DiffusionModule (a.k.a. the MULTI-VERSE of DiffusionModule!) 
DiffusionModule is much cheaper  than the network trunk
than the network trunk  (previous modules).
(previous modules).
→ To improve efficiency,
1.
Network trunk is run only once
2.
(by random rotation, translation and independent noise addition)
3.
DiffusionModule is trained in parallel
•
Loss
◦
Weighted aligned MSE loss (upweighting of nucleotide and ligand atoms)
The famous FAPE loss (from AF2) now disappeared, and authors just used relatively simple MSE loss
◦
smooth LDDT loss (to emphasize local geometry)
◦
bond distance loss (during fine-tuning only)
Confidence Module training with Diffusion “roll-out” 
We need to rollout the DiffusionModule to train AF3 confidence module. Let’s see why.
AF2 Confidence module | AF3 Confidence module | |
heads | - pAE head
- pLDDT head
- experimentally resolved head | - pAE head
- pLDDT head
- experimentally resolved head
- pDE head |
pAE head input | Pair rep (Evoformer output) | Pair rep (mini-Pairformer output - with input feature, single feature, distance between rolled-out coordinate information) |
pLDDT head input | Single rep (Structure module output) | Single rep (mini-Pairformer output - with input feature, pair feature, distance between rolled-out coordinate information) |
Input gradient | Not detached | Detached |
In AF2, confidence module was trained with relatively simple Pair rep and Single rep.
In AF3, confidence module evolved, processing much more information!
•
pDE head (new in AF3): predicts the error in absolute distances between atoms
(this head is also trained with binned classification task)
Other details (such as converting regression and binned classification, pTM & ipTM calculation, …) mostly follows that of AF2.
Distogram prediction
Distogram prediction head is identical to AF2.
Loss
and is 1 for final fine-tuning, 0 for others.
Results
Example structures predicted using AF3
Performance
Accuracy ~ Confidence
Accuracy ~ MSA size
Accuracy ~ Number of seeds
Limitations
Hallucination 
Conformation coverage is limited
Antibody-containing samples require many seeds to obtain high accuracy.
Model outputs do not always respect chirality
Overlapping atoms
Engineering
Dataset composition (TBU)
Training protocols
Model selection
TBU